Two Wide-Angle Imaging Neutral-Atom Spectrometers (TWINS) Mission
The routines in this module can be used to load data from the Two Wide-Angle Imaging Neutral-Atom Spectrometers (TWINS) Mission mission.
Energetic Neutral Atom (ENA) imager
- pyspedas.twins.imager(trange=['2018-11-5', '2018-11-6'], probe='1', datatype='', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, time_clip=False)[source]
This function loads TWINS imager data
- Parameters:
trange (
listofstr) – time range of interest [starttime, endtime] with the format [‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]datatype (
str) – Data type; Valid options: ‘’ Default: ‘’suffix (
str) – The tplot variable names will be given this suffix. Default: ‘’get_support_data (
bool) – If True, data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. Default: Falsevarformat (
str) – The file variable formats to load into tplot. Wildcard character “*” is accepted. Default: ‘’ (all variables loaded)varnames (
listofstr) – List of variable names to load Default: [] (all variables will be loaded)downloadonly (
bool) – Set this flag to download the CDF files, but not load them into tplot variables Default: Falsenotplot (
bool) – Return the data in hash tables instead of creating tplot variables Default: Falseno_update (
bool) – If set, only load data from your local cache Default: Falsetime_clip (
bool) – Time clip the variables to exactly the range specified in the trange keyword Default: False
- Returns:
List of tplot variables created.
- Return type:
Examples
>>> import pyspedas >>> from pytplot import tplot >>> img_vars = pyspedas.twins.imager(trange=['2018-11-5', '2018-11-6']) >>> tplot('smooth_image_val')
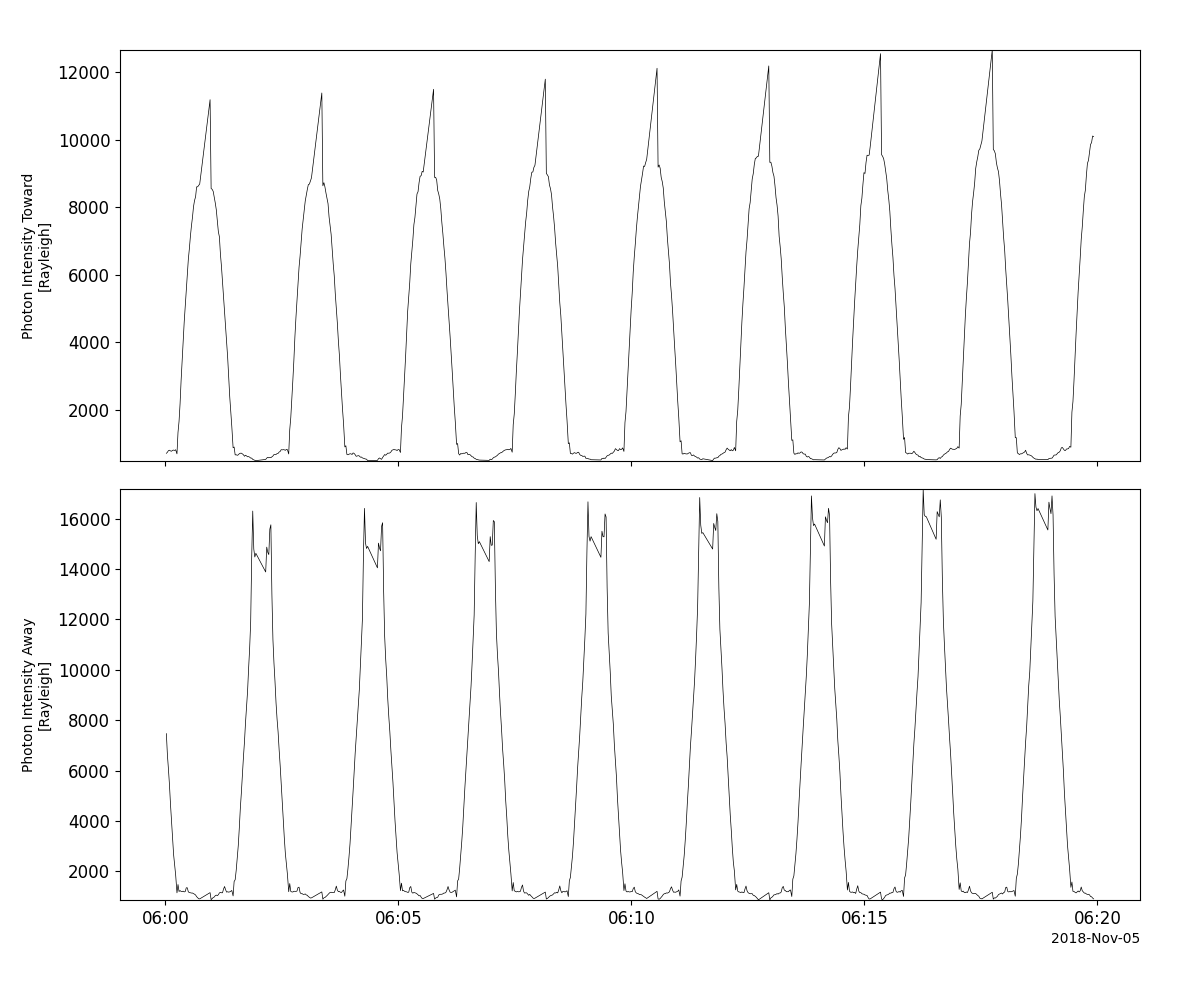
Lyman-alpha Detector (LAD)
- pyspedas.twins.lad(trange=['2018-11-5', '2018-11-6'], probe='1', datatype='', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, time_clip=False)[source]
This function loads data from the LAD instrument
- Parameters:
trange (
listofstr) – time range of interest [starttime, endtime] with the format [‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’] Default: [‘2018-11-5’, ‘2018-11-6’]datatype (
str) – Data type; Valid options: ‘’ Default: ‘’suffix (
str) – The tplot variable names will be given this suffix. Default: ‘’get_support_data (
bool) – If True, data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. Default: Falsevarformat (
str) – The file variable formats to load into tplot. Wildcard character “*” is accepted. Default: ‘’ (all variables loaded)varnames (
listofstr) – List of variable names to load Default: [] (all variables loaded)downloadonly (
bool) – Set this flag to download the CDF files, but not load them into tplot variables Default: Falsenotplot (
bool) – Return the data in hash tables instead of creating tplot variables Default: Falseno_update (
bool) – If set, only load data from your local cache Default: Falsetime_clip (
bool) – Time clip the variables to exactly the range specified in the trange keyword Default: False
- Returns:
List of tplot variables created.
- Return type:
Examples
>>> import pyspedas >>> from pytplot import tplot >>> lad_vars = pyspedas.twins.lad(trange=['2018-11-5/6:00', '2018-11-5/6:20'], time_clip=True) >>> tplot(['lad1_data', 'lad2_data'])
Example
import pyspedas
from pytplot import tplot
lad_vars = pyspedas.twins.lad(trange=['2018-11-5/6:00', '2018-11-5/6:20'], time_clip=True)
tplot(['lad1_data', 'lad2_data'])
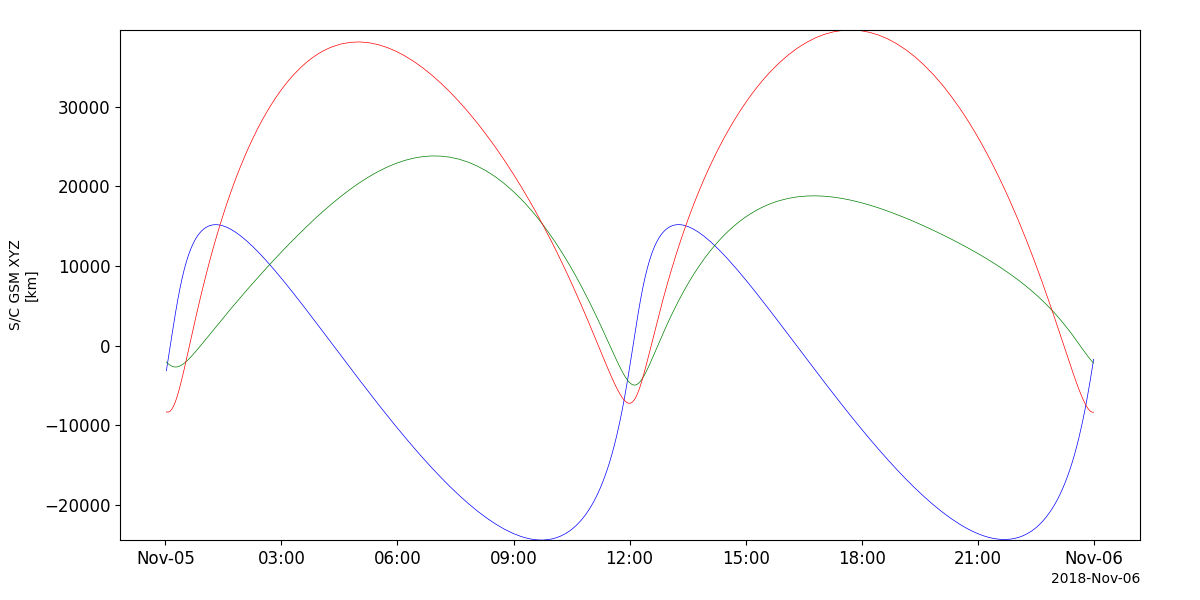
Ephemeris
- pyspedas.twins.ephemeris(trange=['2018-11-5', '2018-11-6'], probe='1', datatype='or', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, time_clip=False)[source]
This function loads TWINS ephemeris data
- Parameters:
trange (
listofstr) – time range of interest [starttime, endtime] with the format [‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’] Default: [‘2018-11-5’, ‘2018-11-6’]probe (
strorlistofstr) – Probe to load. Valid options: ‘1’, ‘2’ Default: ‘1’datatype (
str) – Data type; Valid options: Default: ‘or’suffix (
str) – The tplot variable names will be given this suffix. Default: ‘’get_support_data (
bool) – If True, data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. Default: Falsevarformat (
str) – The file variable formats to load into tplot. Wildcard character “*” is accepted. Default: ‘’ (all variables will be loaded)varnames (
listofstr) – List of variable names to load Default: [] (all variables will be loaded)downloadonly (
bool) – Set this flag to download the CDF files, but not load them into tplot variables Default: Falsenotplot (
bool) – Return the data in hash tables instead of creating tplot variables Default: Falseno_update (
bool) – If set, only load data from your local cache Default: Falsetime_clip (
bool) – Time clip the variables to exactly the range specified in the trange keyword Default: False
- Returns:
List of tplot variables created.
- Return type:
Examples
>>> import pyspedas >>> from pytplot import tplot >>> # Note: variables have the same names for both probes, so only load one at a time >>> ephem_vars = pyspedas.twins.ephemeris(probe=['1'],trange=['2008-04-01','2008-04-02']) >>> tplot('FEQUATORIALGSM')
Example
import pyspedas
from pytplot import tplot
ephem_vars = pyspedas.twins.ephemeris(trange=['2018-11-5', '2018-11-6'])
tplot('FSCGSM')