Arase (ERG)
The routines in this module can be used to load data from the Arase mission.
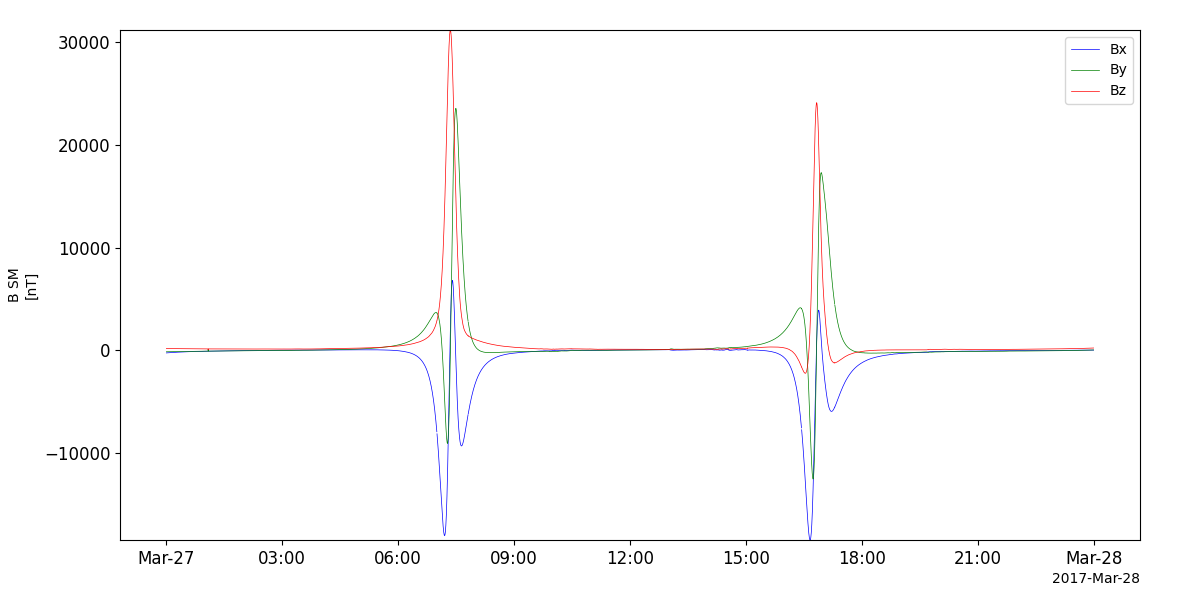
Magnetic Field Experiment (MGF)
- pyspedas.erg.mgf(trange=['2017-03-27', '2017-03-28'], datatype='8sec', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True, coord='dsi', version=None)
This function loads data from the MGF experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
coord – str “sm”, “dsi”, “gse”, “gsm”, “sgi”
version – str Set this value to specify the version of cdf files (such as “v03.03”, “v03.04”, …)
- Returns
List of tplot variables created.
Example
import pyspedas
from pytplot import tplot
pyspedas.erg.mgf(trange=['2017-03-27', '2017-03-28'])
tplot('erg_mgf_l2_mag_8sec_sm')
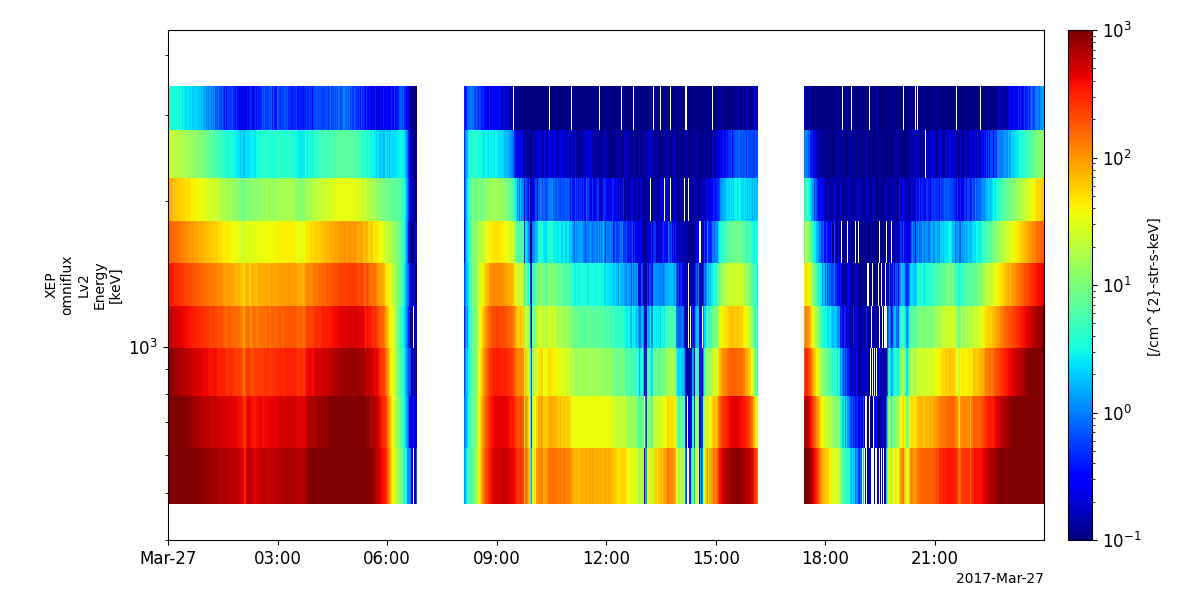
Extremely High-energy electrons (XEP-e)
- pyspedas.erg.xep(trange=['2017-06-01', '2017-06-02'], datatype='omniflux', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True)
This function loads data from the XEP-e experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.xep(trange=['2017-03-27', '2017-03-28'])
tplot('erg_xep_l2_FEDO_SSD')
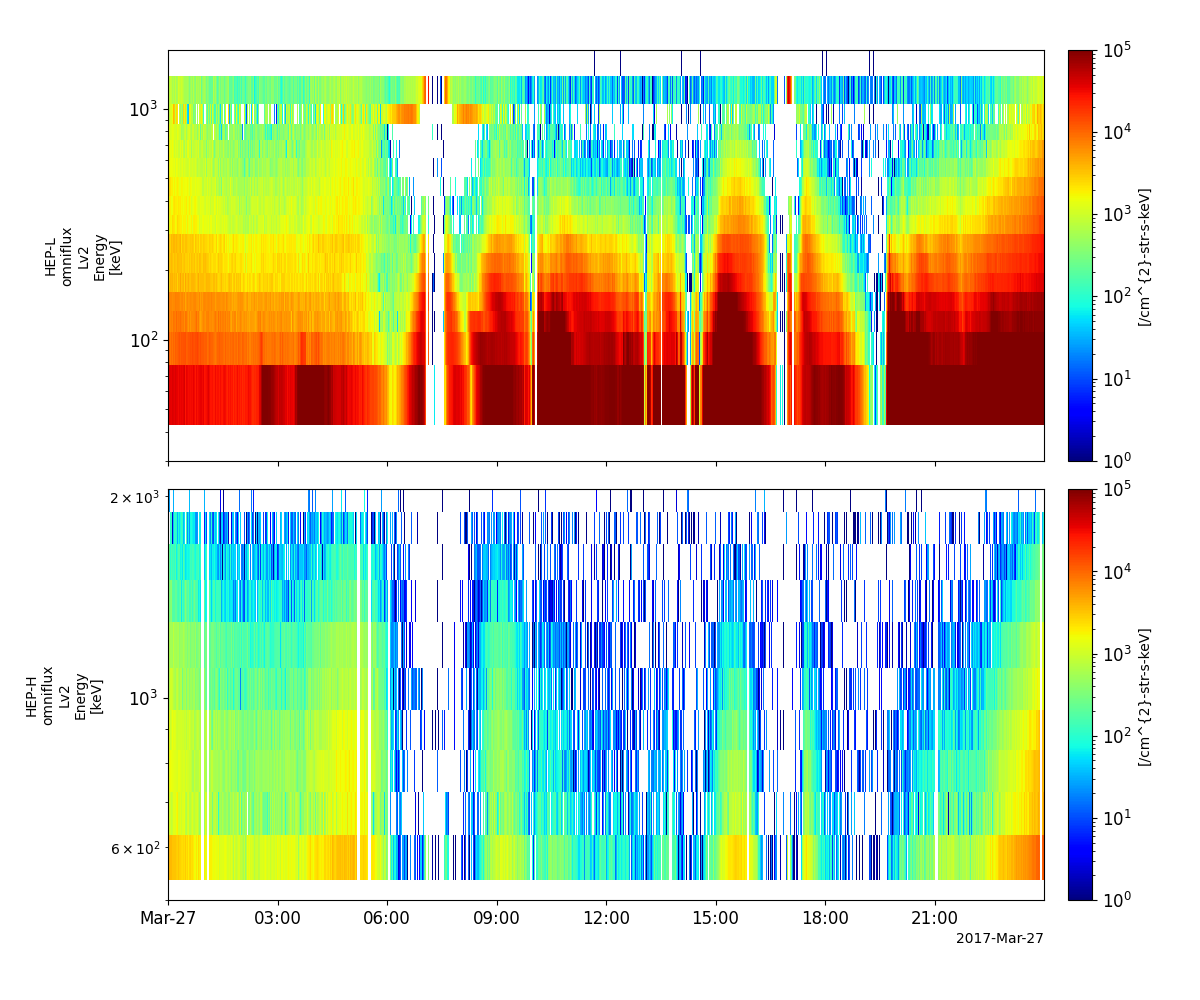
High-energy Particles – electrons (HEP-e)
- pyspedas.erg.hep(trange=['2017-03-27', '2017-03-28'], datatype='omniflux', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True, version=None)
This function loads data from the HEP experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
version – str Set this value to specify the version of cdf files (such as “v01_02”, “v01_03”, …)
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.hep(trange=['2017-03-27', '2017-03-28'])
tplot(['erg_hep_l2_FEDO_L', 'erg_hep_l2_FEDO_H'])
Medium-energy Particles - electrons (MEP-e)
- pyspedas.erg.mepe(trange=['2017-03-27', '2017-03-28'], datatype='omniflux', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True)
This function loads data from the MEP-e experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.mepe(trange=['2017-03-27', '2017-03-28'])
tplot('erg_mepe_l2_omniflux_FEDO')
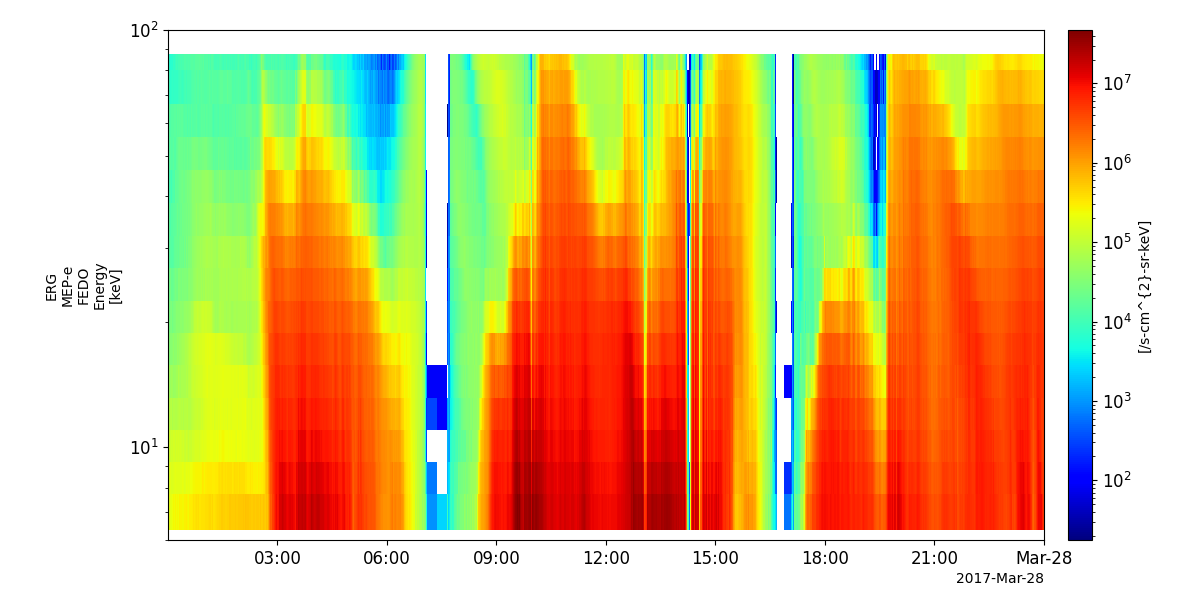
Low-energy Particles – electrons (LEP-e)
- pyspedas.erg.lepe(trange=['2017-04-04', '2017-04-05'], datatype='omniflux', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True, version=None, only_fedu=False, et_diagram=False)
This function loads data from the LEP-e experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
version – str Set this value to specify the version of cdf files (such as “v02_02”)
only_fedu – bool If set, not make erg_lepe_l3_pa_enech_??(??:01,01,..32)_FEDU Tplot Variables
et_diagram – bool If set, make erg_lepe_l3_pa_pabin_??(??:01,01,..16)_FEDU Tplot Variables
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.lepe(trange=['2017-03-27', '2017-03-28'])
tplot('erg_lepe_l2_omniflux_FEDO')
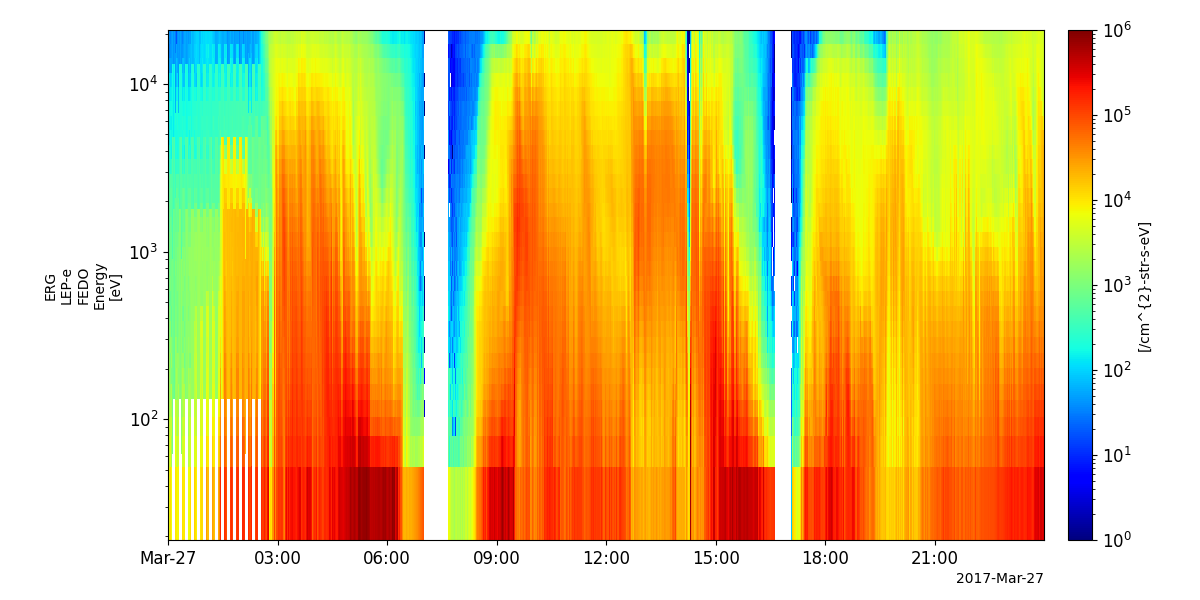
Medium-energy Particles – ion (MEP-i)
- pyspedas.erg.mepi_nml(trange=['2017-03-27', '2017-03-28'], datatype='omniflux', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True)
This function loads data from the MEP-i experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.mepi_nml(trange=['2017-03-27', '2017-03-28'])
tplot('erg_mepi_l2_omniflux_FODO')
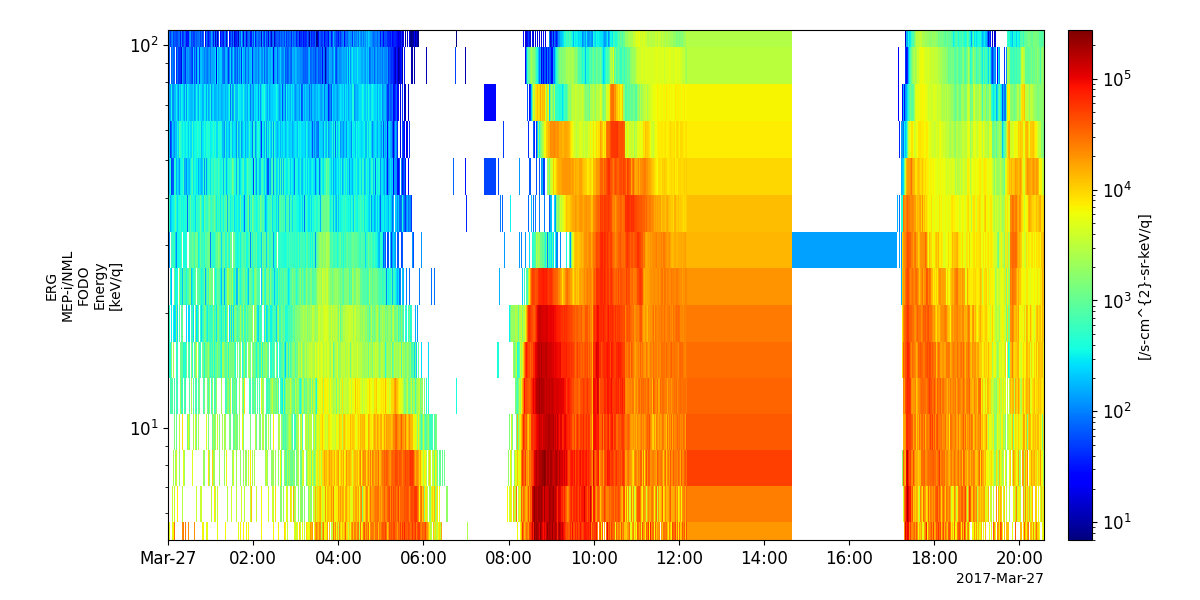
- pyspedas.erg.mepi_tof(trange=['2017-03-27', '2017-03-28'], datatype='flux', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True)
This function loads data from the MEP-i experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
- Returns
List of tplot variables created.
Low-energy Particles – ion (LEP-i)
- pyspedas.erg.lepi(trange=['2017-07-01', '2017-07-02'], datatype='omniflux', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True, version=None)
This function loads data from the LEP-i experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
version – str Set this value to specify the version of cdf files (such as “v03_00”)
- Returns
List of tplot variables created.
Plasma Wave Experiment (PWE)
- pyspedas.erg.pwe_ofa(trange=['2017-04-01', '2017-04-02'], datatype='spec', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True)
This function loads data from the PWE experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.pwe_ofa(trange=['2017-03-27', '2017-03-28'])
tplot('erg_pwe_ofa_l2_spec_E_spectra_132')
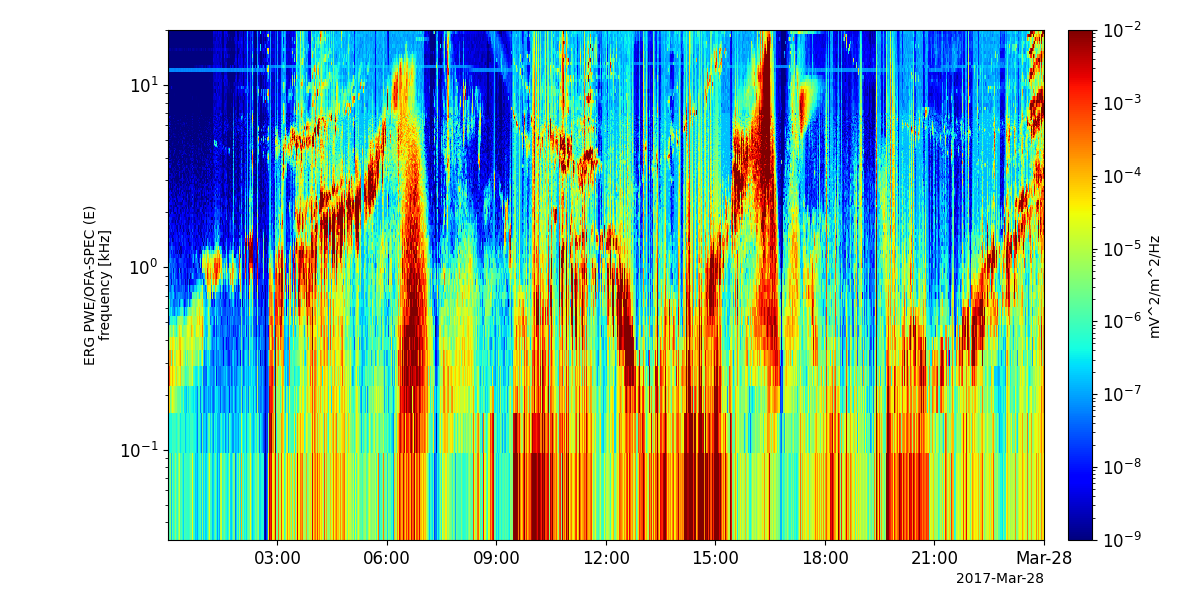
- pyspedas.erg.pwe_hfa(trange=['2017-04-01', '2017-04-02'], datatype='spec', mode='low', level='l2', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True)
This function loads data from the PWE experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.pwe_hfa(trange=['2017-03-27', '2017-03-28'])
tplot('erg_pwe_hfa_l2_low_spectra_esum')
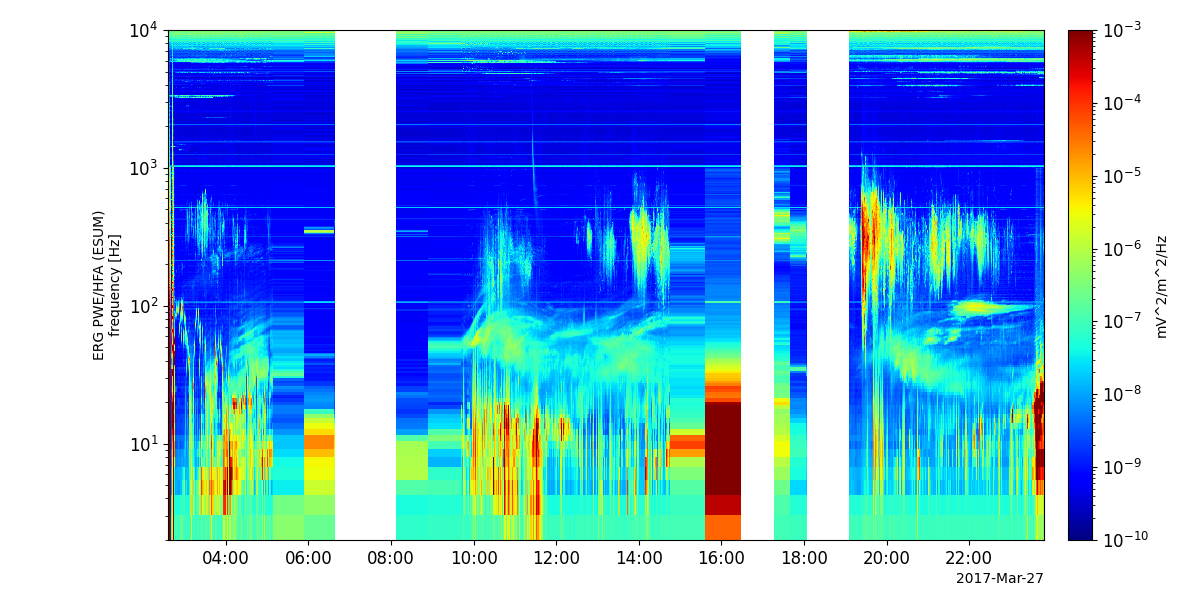
- pyspedas.erg.pwe_efd(trange=['2017-04-01', '2017-04-02'], datatype='E_spin', level='l2', suffix='', coord='dsi', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, ror=True)
This function loads data from the PWE experiment from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
ror – bool If set, print PI info and rules of the road
- Returns
List of tplot variables created.
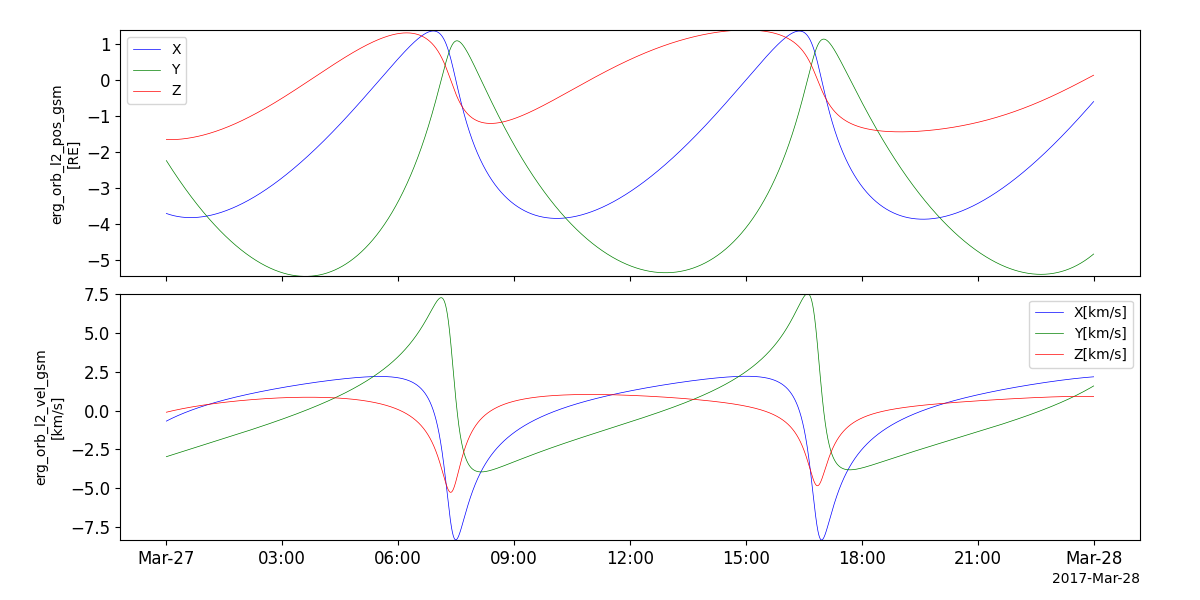
Orbit data
- pyspedas.erg.orb(trange=['2017-03-27', '2017-03-28'], datatype='def', level='l2', model='op', suffix='', get_support_data=False, varformat=None, varnames=[], downloadonly=False, notplot=False, no_update=False, uname=None, passwd=None, time_clip=False, version=None, ror=True)
This function loads orbit data from the Arase mission
- Parameters
trange – list of str time range of interest [starttime, endtime] with the format ‘YYYY-MM-DD’,’YYYY-MM-DD’] or to specify more or less than a day [‘YYYY-MM-DD/hh:mm:ss’,’YYYY-MM-DD/hh:mm:ss’]
datatype – str Data type; Valid options:
level – str Data level; Valid options:
suffix – str The tplot variable names will be given this suffix. By default, no suffix is added.
get_support_data – bool Data with an attribute “VAR_TYPE” with a value of “support_data” will be loaded into tplot. By default, only loads in data with a “VAR_TYPE” attribute of “data”.
varformat – str The file variable formats to load into tplot. Wildcard character “*” is accepted. By default, all variables are loaded in.
varnames – list of str List of variable names to load (if not specified, all data variables are loaded)
downloadonly – bool Set this flag to download the CDF files, but not load them into tplot variables
notplot – bool Return the data in hash tables instead of creating tplot variables
no_update – bool If set, only load data from your local cache
time_clip – bool Time clip the variables to exactly the range specified in the trange keyword
version – str Set this value to specify the version of cdf files (such as “v03”)
- Returns
List of tplot variables created.
import pyspedas
from pytplot import tplot
pyspedas.erg.orb(trange=['2017-03-27', '2017-03-28'])
tplot(['erg_orb_l2_pos_gsm', 'erg_orb_l2_vel_gsm'])